

Audited by an independent third-party, achieving this standard validates our processes in safeguarding customer data.
In accordance with the American Institute of Certified Public Accountants (AICPA) standards for SOC for Service Organizations (SSAE 18)


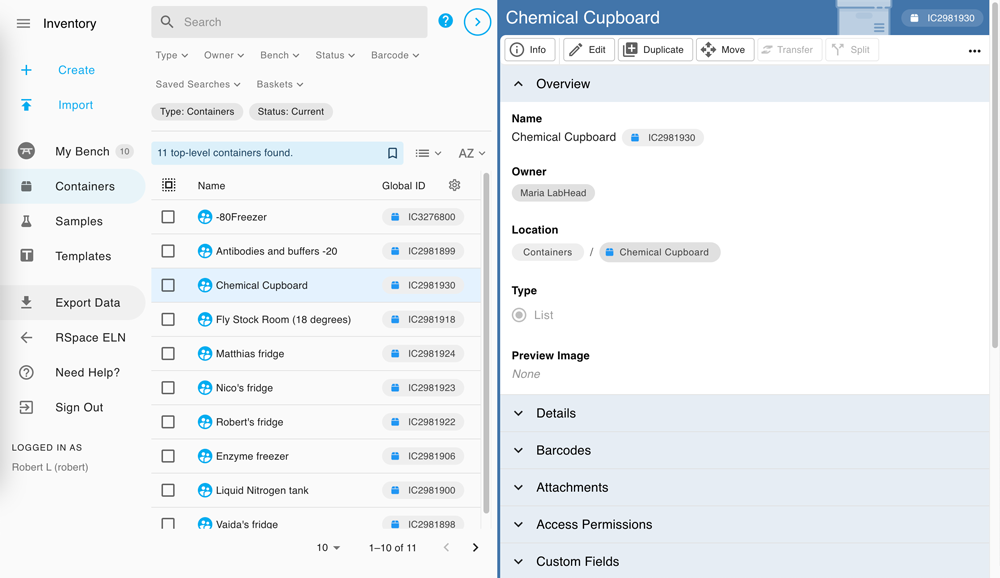
Powerful and smooth sample management, delivered in a flexible and intuitive manner.

We are a DataCite Registered Service Provider, and the first to provide IGSN ID creation and registration services.
✔ Find items easily with a visual, navigable interface
✔ Link your samples and RSpace experiments together with full RSpace integration
✔ Polished features thanks to user-driven development
Learn more about Inventory
Powerful integrations enable you to use all your favourite apps, without hassle.
Learn more about integrations
✔ Private cloud and on-prem deployment options
✔ Customizable data and file management to suit your storage infrastructure and workflows
✔ SSO, hierarchic tiered admin
Under this new policy, all NIH grant applications that collect data will be required to submit a data management and sharing plan (DMSP) with their application. RSpace facilitates compliance through:
✔ Integrations with Dataverse and Figshare, enabling direct deposit of datasets from RSpace to these repositories
✔ Upcoming integrations with Zenodo and Dryad
✔ An integration between RSpace and DMPTool, offering a DMPTool → RSpace → Dataverse/Figshare workflow, enabling full compliance with the NIH Policy: see video below
Everything you'd expect from a modern ELN
See full list of features
✔ Rich content
✔ Folders & notebooks
✔ Chemical structures
✔ Sketches
✔ Snippets & annotations

✔ Share resources with labs
✔ Collaborative editing
✔ Templates
✔ Communities

✔ Link documents & data together for reproducibility
✔ Link to institutional filestores
✔ Link to repositories and archives

✔ Import from Word
✔ Import from protocols.io
✔ Export as HTML, XML, PDF, DOC
✔ Export to an external repository (eg. Dataverse)

✔ 21 CFR 11 compliant
✔ Signing & witnessing
✔ Full audit trail
✔ SSO

✔ Cloud & on-prem options
✔ Custom onboarding
✔ Setup & on-going training and support
✔ SAML2 (Shibboleth) and LDAP (Active Directory) support
✔ System Admin features
A powerful, modern inventory system that's fully integrated with the ELN
See full list of features
✔ Easily configurable sample templates
✔ List, grid, and visual containers
✔ High level of flexibility & customisation
✔ User engagement throughout development
✔ Toggleable views: tree, card, list
✔ Frictionless import flow

✔ Manage sample data from the ELN
✔ Lists of Materials used in experiments for reproducibility
✔ Global IDs of Inventory items for traceability
✔ Directly deposit research & sample data into Figshare & Dataverse

✔ Modern interface
✔ Mobile-first design approach
✔ Location breadcrumbs & parent links for easier navigation
✔ Powerful search-based system
✔ At-a-glance info panels and visual containers
We have designed our software to be easy to use, deploy, and administer.











“Now, the users savor ELN functions like full-text search, revision control, working together on documents, linking files and the possibility to share the ELN across the team and access it from home.”
Read full case study →“When it became clear that the feedback was overwhelmingly positive and users were very happy with the ease of access and quality of support, it was decided that RSpace was the right ELN for CVS.”
Read full case study →

Centre for Cardiovascular Science, The University of Edinburgh
Read case study →
Leibniz Institute on Aging, Fritz Lipmann Institute (FLI)
Read case study →




















✔ All editions include a fully-featured ELN
✔ Unlimited free data storage
✔ Free for individuals and groups
✔ Use it for as long as you like, seamlessly upgrade to Team or Enterprise when you are ready


✔ Private AWS cloud in the supported regional data center closest to you
✔ RSpace Inventory is included
✔ Limited training to support onboarding
✔ Seamlessly upgrade to Enterprise if needed
✔ Private cloud & on-prem deployment options, with setup and on-going support
✔ RSpace Inventory is included
✔ Customizable file management, link to your institutional filestores
✔ SSO support through SAML2 (Shibboleth) and LDAP (Active Directory), hierarchic tiered admin
✔ Custom onboarding, training and user support
✔ Unlimited teaching licenses at no extra cost
